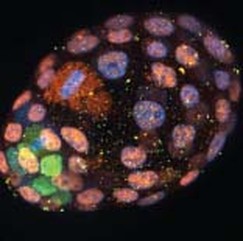
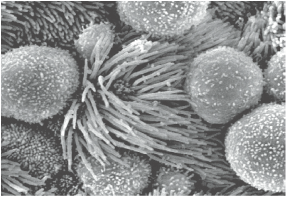
In comparative genomic studies, the number of protein-coding genes clearly fails to correlate with the developmental and pathological complexity in a given organism. Our overall research interest is to investigate the unique biological functions and molecular regulation of various non-coding elements in development and disease. Four major areas of our research are:
Transposons, friends or foes, in mammalian preimplantation development
Transposons in reproductive aging
ncRNAs in development and disease
CRISPR genome editing in disease modeling
Transposons, friends or foes, in mammalian preimplantation development
Transposons in reproductive aging
ncRNAs in development and disease
CRISPR genome editing in disease modeling
Transposons: friends or foes in mammalian preimplantation development~40% of mammalian genome originates from transposons, whose abundance greatly exceeds that of protein-coding genes. While historically viewed as degenerated “parasitic” DNAs, transposons can yield numerous functional elements for their host. These sequences from ancient invasion confer new mechanisms of gene regulation, generate neogene functions, and provide raw material for genome innovation. Mammalian preimplantation embryos constitute one of the best systems to study transposon-host interactions, as 10-20% of their transcriptome results from transposon induction. Using CRISPR genome engineering, genomics, advanced imaging techniques and cell and molecular biology appraoches, we aim to functionally characterize specific transposons to understand how transposon biology are repurposed for preimplantation development.
|
Transposons in disease and aging
While specific transposons are integrated functional components of host biology, most mammalian transposons are potentially detrimental to the host genomes, thus inactivated via degenerative mutations and/or transcriptional/post-transcriptional silencing. Interestingly, aberrant transposon induction has been observed in a variety of pathological conditions, including aging, cancer and infection. We aim to investigate the mechanism underlying transposon dysregulation during cancer and aging, and to elucidate the effects of transposon-host interactions on cellular processes that accelerate disease progression.
While specific transposons are integrated functional components of host biology, most mammalian transposons are potentially detrimental to the host genomes, thus inactivated via degenerative mutations and/or transcriptional/post-transcriptional silencing. Interestingly, aberrant transposon induction has been observed in a variety of pathological conditions, including aging, cancer and infection. We aim to investigate the mechanism underlying transposon dysregulation during cancer and aging, and to elucidate the effects of transposon-host interactions on cellular processes that accelerate disease progression.
|
ncRNAs in development and disease
Non-coding RNAs have emerged as major players for gene regulation in mammalian development and disease. We have characterized the functional importance of miRNAs and long ncRNAs (lncRNAs) in cancer biology, cilia biology and stem cell biology. Our recent effort has been focused on a family of miRNAs that regulate ciliogenesis and mucus production in multiple tissue types. By studying these miRNAs, we have revealed new ciliary ultrastructure, cilia function and cilia dynamics in choroid plexus and in airway epithelia. These findings allow us to elucidate the molecular regulation of cerebral spinal fluid production and circulation, which have profound impact on brain injury and neurodegeneration. |
|
CRISPR genome editing in mice
We recently developed a simple and economic electroporation-based strategy, designated as CRISPR-EZ, to deliver Cas9/sgRNA ribonucleoproteins (RNPs) into mouse zygotes with 100% efficiency. This technology bypasses microinjection, enabling highly efficient and high-throughput genome editing in vivo, with a significant improvement in embryo viability. We are now building up upon this technology to establish new CRISPR editing system to study subcellular localization of endogenous proteins and RNAs. |
 |